pipefunc 🕸️
🎄🎁 Advent of Open Source – Day 20/24: A Python package to streamline scientific computations with minimal boilerplate.
(See my intro post)
Of all the projects I’m sharing this month, this one that started as a passion project excites me the most!
📖 Origin Story #
Sometimes the best projects are born from the most unexpected moments. More than 1½ year ago, while on parental leave with my twin boys, I found myself with small pockets of time during naps to work on a problem that had been bothering me for years: the tedious bookkeeping required in complex computational workflows. Every scientific computation project seemed to reinvent the same patterns - managing function dependencies, parameter sweeps, result caching, parallelization, and a lot of boilerplate to combine the resulting data. I wanted something that would let scientists focus on their science, not on pipeline management.
🔧 Technical Highlights #
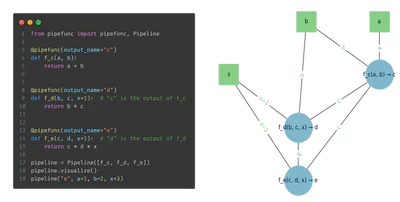
- Automatic DAG construction via very simple and lightweight syntax
- N-dimensional parameter sweeps with automatic parallelization
- Visual pipeline representation using NetworkX
- Resource profiling (CPU, memory, time)
- Type validation between pipeline stages
- Ultra-fast: only 15 µs overhead per function
- Flexible caching strategies (memory, disk, cloud)
- Integration with scientific computing tools:
📊 Impact #
- 230 GitHub stars
- 700+ tests with 100% coverage
- Fully typed codebase
- Comprehensive documentation
- Tested on real workflow on SLURM cluster
- Covered on Pycoder’s Weekly with >100k subscribers
🎯 Challenges and Solutions #
- Balancing simplicity with power
- Making complex workflows intuitive
- Handling distributed computing edge cases
- Ensuring type safety across the pipeline
- Optimizing performance without sacrificing features
💡 Lessons Learned #
- Sometimes the best time to code is during baby naps
- Complex problems can have elegant solutions
- Scientific computing needs better tooling
- Good abstractions make hard things easy
- Type hints and tests prevent headaches
🔮 Future Plans #
The journey is far from over. Plans include:
- Enhanced cloud computing support
- More interactive visualization options
- Interactive pipeline debugging tools
- Expanded parameter sweep capabilities
Want to simplify your computational workflows? Check out pipefunc on GitHub or read the documentation!
#OpenSource #Python #DataScience #ScientificComputing #Programming